Abstract
The electronic health record allows the assimilation of large amounts of clinical and laboratory data. Big data describes the analysis of large data sets using computational modeling to reveal patterns, trends, and associations. How can big data be used to predict ventilator discontinuation or impending compromise, and how can it be incorporated into the clinical workflow? This article will serve 2 purposes. First, a general overview is provided for the layperson and introduces key concepts, definitions, best practices, and things to watch out for when reading a paper that incorporates machine learning. Second, recent publications at the intersection of big data, machine learning, and mechanical ventilation are presented.
Introduction
Artificial intelligence and machine learning are tailored to identify complex, nonlinear relationships in large volumes of data. The typical patient admitted to the ICU is likely to be connected to a physiologic monitor, a mechanical ventilator, and various medication infusion pumps and to receive a number of tests, all of which generate a vast amount of data. These data, when combined with information obtained by bedside clinicians and entered into the electronic health record, make the ICU an extremely data-rich environment. However, utilizing these data to their fullest extent is difficult. Further, many aspects of care in the ICU are not informed by high-quality, large, randomized controlled trials (RCTs), and practice variability among providers can be high.1,2 The sheer volume of data, the complexity of individual patients, and variability in practice represent an opportunity to apply big data techniques, artificial intelligence, and machine learning to provide solutions to important clinical problems. Such solutions may involve predicting clinical deterioration, automated identification of patients who are ready to extubate, early warning signs of important conditions such as sepsis to permit early intervention, and providing decision support to clinicians to optimize mechanical ventilation.3
Types of Clinical Data
For a subject admitted to the ICU and receiving mechanical ventilation, there are disparate data types that must be considered when storing, accessing, and deriving information from the data. Structured data elements are those that document patient information using controlled vocabulary rather than a text narrative or other unstructured means.4 An example would be a flow sheet row in the medical record where a clinician records breathing frequency. Data from drop-down lists, such as those utilized to document breath sounds, or capnographic data are also examples of structured data. In most cases, structured data are preferred because the data are relatively easy to query, manipulate, and process.
The term “unstructured data” has gained popularity in recent years and goes hand-in-hand with big data. These are data that cannot be readily mapped to a specific field with known characteristics. Unstructured data often are stored without a clear purpose for later use, or it is very difficult or impossible to impose a structure on it. Examples include clinical notes in which a clinician can enter free text, most imaging information, and a few other sources that typically come from outside the electronic health record.
There is a need to integrate these data types to achieve the maximum clinical, quality, and business value.5-7 There are efforts in the field to adopt standards that facilitate the flow of data. One such effort is clinical document architecture, which is an important standard in the United States that seeks to incrementally structure data and provide interoperability.8 As these standards are implemented, the hope is that individual institutions, departments, and teams will be able to more readily access and extract value from these data.
Data Collection and Warehousing
Data collection at the bedside is particularly daunting, especially because a single subject can be connected to several devices that output different types of data at different frequencies. First and foremost, the admission-discharge transfer feed, which contains important patient identifiers, is used to associate the patient with the bed space, medical record, and other data sources. A data warehouse is a central location for all data that an enterprise or hospital collects.9 These data can include those stored directly in the electronic health record (including the medication administration record, physiologic data, treatments, notes), data incorporated into the admission-discharge transfer, continuous physiologic data from medical devices (eg, monitors, mechanical ventilators, radiologic or ultrasound imaging), billing, and other sources. Increasingly, institutions are implementing data architecture that streamlines the collection of data from multiple locations to a central repository for patient care, quality improvement, and research.
Data Mining and Data Science
As the field of artificial intelligence has progressed over the past 30 y, data mining has been of prominent interest among researchers in the field.10 Data science is the science of extracting useful information from data sets, and it spans several disciplines, including statistics, data management, artificial intelligence, machine learning, and pattern recognition among others.11 A researcher engaged in data mining will address data collection, storage and retrieval, data cleaning, data reduction, visualization, algorithm development, machine learning, and statistical analysis, and will also need to balance statistical and computational issues.12 Some common data science terminology is provided in Table 1.
Key Concepts and Common Terminology
The role of the data scientist is one that spans various industries and all levels of enterprise decision making. A data scientist works at the intersection of substantive expertise (domain knowledge), mathematics and ap-plied statistics, machine learning, and coding skills (https://berkeleysciencereview.com/2013/07/how-to-become-a-data-scientist-before-you-graduate, Accessed August 22, 2019). On one hand, all scientists deal with data, and one could bestow the title of data scientist upon anyone dealing with data and statistics. However, for our purposes, this is referred to as traditional research. As more data are collected, knowledge of machine learning and coding skills are required to extract the maximum value from patient data. It is in the best interest of the field of medicine to have people with extensive substantive expertise (eg, respiratory therapists, doctors, nurses, and other clinicians) gain the skills needed to become data scientists. One of the issues facing a data scientist with little knowledge of a given field is solving problems that either don’t exist or are obvious to clinicians in the field. Data scientists with keen awareness of clinical practice, especially as it pertains to mechanical ventilation, will be in an excellent position to achieve insight into pathophysiology and development of useful decision-support tools that will have a clinical impact.
Big Data
The origin of the term “big data” is not clear. The term itself is very broad, is debated loudly, and often is not helpful in conversation. The most clear and helpful definition is that ‘big data’ is a high-volume, high-velocity, and high-variety information asset that demands cost-effective, innovative forms of information processing for enhanced insight and decision making. A part of the definition includes the 3 Vs: volume (quantity of data), velocity (rate at which data are generated and collected), and variety (various types of data and sources). In general, big data are the ocean in which a variety of specific data tasks are found.
Artificial Intelligence
The definition of artificial intelligence can be controversial depending on the applied domain (eg, computer science, data science, statistics, science fiction). Alan Turing, English mathematician and widely recognized father of artificial intelligence, devised what is known as the Turing Test of computer intelligence.13 Turing proposed that if a computer could mimic human behavior, and in so doing fool a human into believing they were interacting with a human, that computer could be defined as possessing intelligence. More broadly, artificial intelligence is defined as the branch of computer science dealing with the simulation of intelligent behavior in computers and the capability of a machine to imitate intelligent human behavior (https://www.britannica.com/technology/artificial-intelligence, Ac-cessed August 22, 2019).
Machine Learning
Machine learning is a subset of artificial intelligence that provides a mechanism for a computer algorithm to learn from data and improve through experience without being explicitly programmed.14 This is particularly useful in the context of medicine because it may not be completely necessary to explain (and program) all of the variance in a given subject to get a reasonable approximation of system performance or subject health in an effort to provide individualized care. In general, there are 2 principal types of learning utilized within machine learning that a researcher can employ to extract knowledge from a given dataset: supervised learning and unsupervised learning.
Supervised Learning.
Supervised learning describes a problem where a discrete outcome is known a priori. Techniques within supervised learning utilize a labeled dataset that has 2 components: (1) a set of inputs (typically a vector of data containing either continuous variables, categorical variables, or a combination of both) and (2) a target or outcome. This is collectively referred to as the training data and is used to train the machine learning algorithm.
Unsupervised Learning.
Unsupervised learning is the machine learning task of uncovering hidden structure or relationships within an unlabeled dataset.15 An unlabeled dataset is one where no target or outcome exists. An example of unsupervised learning is the application of a cluster-analysis technique to detect distinct phenotypes of subjects with a common clinical diagnosis.16 In this case, this information may reveal differences in clinical characteristics and prognosis for a given diagnosis, such as bronchiectasis.
Semisupervised Learning.
A third type of learning is called semisupervised learning. Semisupervised learning occurs when input data are a mixture of both labeled and unlabeled cases. In this case, the prediction problem is composed of identifying the organizational structure of the data as well as the predictions. Often, these algorithms extend the functionality of other methods.
Some examples of machine learning algorithms seen in the medical literature included tree-based methods, discriminant analysis, regression models (multiple and logistic), support vector machine, k nearest neighbor, and neural networks. An important aspect of applying machine learning in health care is the interpretability of the algorithm. Tree-based algorithms, discriminate analysis (depending upon the discriminant function), and support vector machines (depending on the function used) are generally preferred for their improved interpretability.17 Support vector machines and discriminate analysis have been applied to research problems in the ICU and during mechanical ventilation.18-20 Selecting a machine learning algorithm should be a balance of overall performance (accuracy), duration of time needed for training, and interpretability.17,21
Classification and Regression
In general, machine learning is utilized to make a prediction or observation about a variable. Machine learning can be divided broadly into regression problems and classification problems.11 A regression problem is one where the output is a continuous variable in either space or time. During mechanical ventilation, it may be desirable to predict  and
and 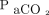 of a patient continuously during or after a clinical intervention. In the context of a regression, an algorithm could be constructed to predict the
of a patient continuously during or after a clinical intervention. In the context of a regression, an algorithm could be constructed to predict the 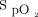 and
and  of a patient at any point in time; for example, if PEEP is increased,
of a patient at any point in time; for example, if PEEP is increased, 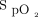 and
and 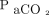 are predicted to be 93% and 42 mm Hg, respectively. On the other hand, a classification approach may involve simply categorizing a PEEP change as generally good or generally bad (either a positive or negative response). It is important to note that many problems can be posed as either a regression problem or a classification problem (each with benefits and drawbacks). It is essential to understand the clinical problem, the objective of the decision-support tool you are developing, and the statistical performance of the algorithm when deciding which methodology is most appropriate.
are predicted to be 93% and 42 mm Hg, respectively. On the other hand, a classification approach may involve simply categorizing a PEEP change as generally good or generally bad (either a positive or negative response). It is important to note that many problems can be posed as either a regression problem or a classification problem (each with benefits and drawbacks). It is essential to understand the clinical problem, the objective of the decision-support tool you are developing, and the statistical performance of the algorithm when deciding which methodology is most appropriate.
Need for RCTs
Big data and applied machine learning will not completely replace the need for RCTs. A well-designed and well-conducted RCT has the advantage of making causal inferences based on the random assignment of subjects to different treatment groups. In any investigation, there are a number of known and unknown factors that can have an impact on outcomes outside of the treatment assignment. So long as these factors are distributed randomly between the treatment and control groups, statistical differences in outcomes can be attributed to the treatment. Suppose one has access to a large amount of clinical data (ie, big data). Could a study be designed and applied retrospectively to detect treatment causation? One such retrospective design that relies upon big data is the case-matched controlled study. The degree to which cases are matched depends largely on the dimensionality of the data (ie, the number of variables describing each subject or case): the more data available to match patients, the higher the probability of the findings being valid. Certainly, a high-quality retrospective case-matched controlled study will provide important details, but we caution against equating retrospective big data studies as equal to an RCT. That said, there are a number of disadvantages to an RCT: high cost, study population is often too narrow to make broad conclusions, and the time between study commencement and translation to clinical practice is very long. Therefore, a role for a fast, low-cost, and effective investigation tool is needed to bridge the gap and to help inform best practice and refine hypotheses for RCTs that may have a higher probability of positive results.
Best Practice (What to Look Out for When Reading a Paper That Includes Machine Learning)
It’s unnecessary for the average clinician or respiratory researcher to be an expert in the fields of big data, artificial intelligence, and machine learning. However, as these methods are more frequently applied in the literature, it is helpful to be familiar with some best practices that can help distinguish between good and poor study design. In general, the goal of any prediction model is to obtain the best performance that permits good generalizability. A handful of issues to watch out for are discussed below.
Great Data (and an Outcome That You Care About).
Like any high-quality scientific work, one must begin with high-quality data. For a database to be sufficient and to be deemed high-quality, several elements are required: accurate data, sufficient size (ie, number of cases or subjects), sufficient dimensions (ie, number of predictor variables), labeled cases (ie, the outcome measure, which can be continuous, binary, or categorical).
Of particular interest is selecting an outcome measure that is clinically meaningful. One of the pitfalls that can be observed in the field of applied machine learning in health care is that some studies are conducted with an irrelevant clinical target, are based on data that are not routinely available to others, or do not meet acceptable sensitivity and specificity to justify clinical implementation. Put another way, it’s very possible to spend a lot of time, energy, and resources solving a problem that either does not exist (ie, isn’t important enough to proceed to clinical utility) or does not offer a significant improvement over existing practices or existing clinician proficiency.
All About the Numbers.
Machines require a lot of data to “learn,” but humans do not. A respiratory therapy student can have a bedside teacher explain the relationship between dead-space ventilation and the 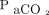 /
/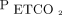 gradient a single time (or perhaps a handful of times) to understand the principal and foresee possible clinical implications. Machines need to see hundreds, thousands, or millions of cases to identify the concept. Therefore, any project in the area of machine learning should have a relatively large number of cases. This number can range anywhere from a few hundred cases to thousands of cases to many more. The higher the number of cases, the higher the probability that the model can identify a process that actually exists.
gradient a single time (or perhaps a handful of times) to understand the principal and foresee possible clinical implications. Machines need to see hundreds, thousands, or millions of cases to identify the concept. Therefore, any project in the area of machine learning should have a relatively large number of cases. This number can range anywhere from a few hundred cases to thousands of cases to many more. The higher the number of cases, the higher the probability that the model can identify a process that actually exists.
Testing (Not Just for Professional Cyclists).
The purpose of testing is to assess the ability of a model to make predictions on data that were not used to train the model.11 The easiest type of testing to conceptualize is a holdout scheme. After data have been compiled, a specific portion of the cases will be randomly assigned to a training data set and a testing data set. Typical ratios of training and testing cases are anywhere from 50/50 to 80/20 (ie, 80% of cases are used to train the model and 20% are used solely to test the model and to compute performance statistics). Other acceptable methods of testing include some type of cross-validation. Cross-validation is typically performed 5–10 times (ie, 5-fold cross-validation). At each step, a portion of the data are used to train the model, and the other portions are used to compile performance statistics.22 An example of a 5-fold cross-validation is depicted in Figure 1.
Illustration of 5-fold cross-validation. In the first iteration, a portion of the data (one fifth of all cases) is assigned to testing, with the remaining data being available for training the machine learning model. In the second iteration, a different portion of the data (but still one fifth of all data) is assigned to testing. This process continues until all cases have been used for training and for testing. Summary statistics are compiled from each iteration, and the final results that are communicated in a paper reflect the average results from each iteration.
The training and testing of machine learning models is also necessarily based on data from the past. This fact is not necessarily bad, but one must be mindful that these data may include information about medical practices that are not ideal, or the cohort may be affected by selection bias. In general, past performance cannot guarantee future success. As was mentioned earlier in this review, it is unlikely that the field of data science and machine learning in health care will completely replace RCTs. However, in many cases, pragmatism (ie, not enough money or time) will force us to conduct studies on retrospective data and not go through with an RCT before implementing something new in clinical practice. On the other hand, a reasonable path forward in many other cases would be to compile historical data, ensure its quality, apply appropriate statistical and machine learning techniques to extract an accurate model, and test this model prospectively in a well-designed, prospective clinical study.
Overfitting (Some Things Just Don’t Fit Anymore).
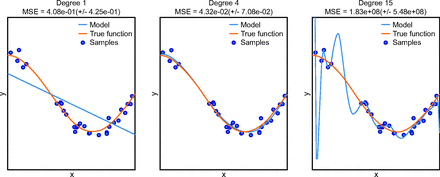
Over-fitting refers to a model that produces predictions that too closely or exactly fit a particular data set and therefore fails to generalize performance to other data.23 Overfitting can occur if careful attention is not paid to study design, especially as it pertains to training and testing scheme (Fig. 2).24
Example of overfitting. The true function is depicted by an orange line, and samples are shown as circles. The 3 panels show the result of adding degrees to a polynomial prediction model (blue line). In the left panel, a polynomial of 1 degree (ie, linear) is shown. This does not adequately describe the sample data and is said to be underfit. In the right panel, a polynomial of 15 degrees is shown. This model is exactly fit to each sample in the training set, and one may be prone to believe that, because the model exactly fits the training samples, it must be the best model; however, this is an example overfitting because the model does not match the true function, which is shown by the orange line. If research offered no out-of-sample validation, the ability of the model to make future predictions is unknown and the model could be overfit, which may make future performance quite bad. In the middle panel, a polynomial of 4 degrees is applied. The model does not exactly predict each sample but the general relationship between the predictor variable (x) and outcome (y) is adequately described, and the accuracy of future predictions made on data that are unknown at this time would be quite good. From Reference 24, with permission. MSE = mean squared error.
The curse of dimensionality refers to a number of adverse phenomena that occur when analyzing data that have a high dimensional space; dimensions in this context are variables or inputs). Simply put, the higher the number of inputs, the lower the accuracy of the model to make future predictions. Methods that can mitigate the curse of dimensionality include feature selection and dimensionality reduction. Both of these methods seek to reduce the overall number of features such that the model yields both good prediction performance and generalizability.
Model Selection (Choose Wisely.).
Machine learning is not a goal but a tool. One should not apply a neural network when a logistic regression model will do the job. In the medical literature, it is desirable to understand the relationship between individual variables and the outcome of interest for important reasons: understanding underlying pathophysiology, identifying mechanisms of action, and hypothesizing possible clinical interventions. That said, the simplest and easiest to interpret model is always preferred if there is not a significant difference in performance compared to a more complicated model. Neural networks offer no explanatory power. Other models like decision trees or support vector machines provide the capability to examine relationships among predictor variables and the outcome of interest. It is important, therefore, when reading a paper that applies a black box model (ie, one which cannot be examined, like a neural network) with no rationale or benchmarking against other interpretable models.
Mechanical Ventilation
Mechanical ventilation is an essential clinical intervention applied to > 800,000 patients/y and to ∼40% of patients in the ICU.25 More than any other single device in the ICU, the mechanical ventilator offers a large amount of data in the form of settings, measured parameters, alarm status, and waveform data. When these data are coupled with the fact that patients who receive mechanical ventilation account for a disproportionate amount of health care spending, a large number of opportunities exist in this area to improve the efficacy, efficiency, and safety of care by implementing machine learning.
ARDS
ARDS is the result of a number of disparate risk factors that occur either locally or systemically.26 ARDS occurs in 10.4% of ICU admissions and is commonly underrecognized, undertreated, and associated with a high mortality rate.27,28 However, identifying patients who are likely to develop ARDS in the ICU remains an important challenge because timely diagnosis and appropriate treatment can improve outcomes.
Afshar et al29 sought to develop a computable phenotype for ARDS using natural language processing and machine learning. A computable phenotype is a method used to define a condition, disease, clinical event, or other patient characteristic using only data available to and processed by a computer.30 The authors reported that, combined with their best natural language processing model, accuracy of the identified computable phenotype was 83% (95% CI 58.3–76.3). This result was superior to the benchmark algorithm tested, but it still leaves much room for improvement. Certainly, future studies should not be limited to only text data but should include physiologic data such as blood gas values, oxygen saturation, 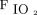 , and other elements of ventilation. The work of Afshar et al,29 however, is an important step forward because it seeks to determine objective and reproducible methods of diagnosing ARDS that depend less on human factors.
, and other elements of ventilation. The work of Afshar et al,29 however, is an important step forward because it seeks to determine objective and reproducible methods of diagnosing ARDS that depend less on human factors.
Apostolova et al31 sought to predict the development of ARDS by combining structured and unstructured data. Their structured data included available diagnosis codes, physiologic monitor data, and laboratory data; unstructured data included clinical notes. The salient feature of this work is the combination of structured and unstructured data. The authors applied a deep learning approach to build a patient context vector that contains summary information about the patient’s current condition. The patient context vectors could then be combined with the structured data (eg, labs, vitals, etc.) and a prediction model was trained to predict ARDS. The top model was a gradient-boosted machine that demonstrated an area under the receiver operating characteristic curve of 0.93. The top 5 features in the model were minimum tidal volume, Glasgow coma score, respiratory rate, 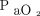 , and age.31
, and age.31
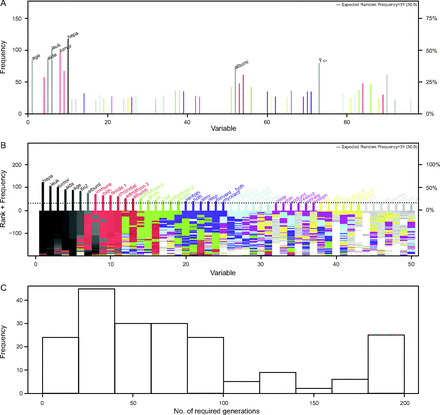
In a secondary analysis of 2 multi-center RCTs from 44 hospitals, Zhang32 conducted a study in the same area. However, rather than predict development of ARDS, the author sought to predict mortality following diagnosis of ARDS and to provide risk stratification in an effort to help clinicians choose appropriate treatment. A genetic algorithm was utilized to identify features of importance in the available data, and a neural network was trained to perform the prediction. Although used in other fields, genetic algorithms have not been frequently applied to clinical data. A genetic algorithm is based on the concept of natural selection and performs an adaptive heuristic search.33 From 88 candidate variables (including demographics, details of the admission, laboratory data, physiologic data and information from the mechanical ventilator), 7 variables were identified to be most important: age, history of acquired immunodeficiency syndrome, leukemia, metastatic tumor, hepatic failure, lowest albumin, and  . A graphical representation of the genetic algorithm is depicted in Figure 3. Indeed, any reasonably informed clinician would likely generate a similar list because each of these factors could be independently associated with increased mortality. Nonetheless, the area under the receiver operating characteristic curve for the neural network was 0.82 (95% CI 0.75–0.89), which outperformed the APACHE III score of 0.67 (95% CI 0.59–0.74). Although interesting, the ability of the algorithm to be extrapolated to other populations remains an open question. The methods by which important variables were identified nonetheless remain an important contribution of this work by Zhang.32
. A graphical representation of the genetic algorithm is depicted in Figure 3. Indeed, any reasonably informed clinician would likely generate a similar list because each of these factors could be independently associated with increased mortality. Nonetheless, the area under the receiver operating characteristic curve for the neural network was 0.82 (95% CI 0.75–0.89), which outperformed the APACHE III score of 0.67 (95% CI 0.59–0.74). Although interesting, the ability of the algorithm to be extrapolated to other populations remains an open question. The methods by which important variables were identified nonetheless remain an important contribution of this work by Zhang.32
Results of a genetic algorithm search. A: shows the frequency of each “gene” (ie, clinical variable) presented in stored “chromosomes” (ie, a combination of clinical variables). The top 50 variables are colored, and the top 7 variables were named. B: displays the stability of the rank of the top 50 variables; the top 4 variables appeared to stabilized faster than others. C: shows the distribution of the number of generations required for an evolution to achieve the fitness goal. If an evolution epoch cannot reach the fitness goal of area under the receiver operating characteristic curve = 0.77, the iteration is considered as “no solution” and it is stopped. The training sample was split into the training and test sets in a 2:1 ratio. AIDS = acquired immunodeficiency syndrome; tumor = metastatic tumor; leuk = leukemia; hepa = hepatic failure; bilih = highest bilirubin; albuml = lowest albumin; immune = immunodeficiency; hcth = highest value of hematocrit; reside = residence prior to admission; admitfrom = admission source; gluch = highest glucose; pip = peak inspiratory pressure on day 0; resp = breathing frequency on day 0; sodiumh = highest sodium value. From Reference 33, with permission.
Toward Precision-Guided Recruitment Maneuvers
Despite sound physiologic rationale, recruitment maneuvers are not routinely recommended for patients with ARDS.34,35 Although recruitment has been shown to be detrimental among all patients with ARDS, there may be a small subgroup of patients who respond positively and would receive an important benefit. Identifying this subgroup, however, has been difficult. Zampieri et al36 conducted a post hoc analysis to examine whether a portion of ARDS patients could benefit from early alveolar recruitment. Because traditional methods of subgroup analysis failed to identify a subgroup of subjects for whom recruitment could be beneficial, the authors applied a machine learning method of clustering known as k-mean clustering. A k-mean clustering algorithm is a type of unsupervised learning that seeks to identify groups in the data. The variable k refers to the number of groups identified. The clustering method identified a cluster that exhibited an association between recruitment maneuvers and PEEP titration with increased probability of harm. In Figure 4, the 3 clusters that were identified with the machine learning method are depicted. Importantly, this method provides a probability distribution of each cluster and whether they are more likely to benefit from standard ARDSNet ventilation without recruitment or care with the addition of alveolar recruitment. Through a precision medicine approach, the results suggest that a very small proportion of ARDS patients are likely to benefit from recruitment maneuvers (Fig. 4, see subjects in cluster 2). Indeed, this method should be applied to other areas of mechanical ventilation to identify subgroups and to provide improved decision support to bedside clinicians.
Results of Bayesian heterogeneity in treatment effect. Posterior probability distribution of the Alveolar Recruitment for ARDS Trial (ART) treatment effect [log(RR)] in each cluster. The table in the upper left corner contains the probability that the relative risk for mortality using the ART treatment is < 0 (ie, a relative risk < 1, suggestive of a protective effect of the ART treatment) for each cluster. From Reference 36, with permission.
Automated Patient–Ventilator Asynchrony Detection
Patient–ventilator asynchrony is associated with increased risk of mortality and increased duration of mechanical ventilation.37 Sottile et al38 developed a collection of machine learning models that identify the various types of asynchrony: double-trigger, flow limit/starvation, premature breath termination, and ineffective trigger. The authors applied random forests, Gaussian naïve-Bayes, and ADABOOST algorithms to each type of dyssynchronous breath.39 Overall, area under the receiver operating characteristic curve ranged from 0.954 to 0.972 and exhibited very good sensitivity and specificity. Should this performance be observed prospectively, it may represent an important alert that will enable clinicians to adjust mechanical ventilation or other aspects of treatment and obviate the perceived increased risk of mortality and duration of ventilation. Certainly, further work in this area is needed to reproduce these results in a broader patient population, including both restrictive and obstructive lung disease and across patient types (ie, neonatal, pediatric, and adult). However, these methods may be coming to a ventilator near you sooner than one may suspect.
Prolonged Mechanical Ventilation and Tracheostomy
Predicting need for prolonged mechanical ventilation could aid in tracheostomy tube placement, weaning strategy, and disposition planning. Parreco et al40 used data from the Multi-parameter Intelligent Monitoring in In-tensive Care III (MIMIC III) database to build a classifier for predicting prolonged mechanical ventilation (ie, > 7 d) and tracheostomy tube placement. A gradient-boosted decision tree model was trained using available demographic, diagnostic, laboratory, and other clinical data. Overall, performance of the classifiers were generally good, and the area under the receiver operating characteristic curve was 0.852 ± 0.017 and 0.869 ± 0.015 for prolonged mechanical ventilation and tracheostomy, respectively. However, in both cases, sensitivity of the classifiers was low at 47.8% and 26.8% for prolonged ventilation and tracheostomy, respectively. As such, the clinical utility of the present model is unclear. Additionally, it is unclear whether these results outperform a prediction by a bedside clinician. For instance, if a patient presents to the ICU with significant pulmonary system dysfunction (defined with something like a LODS score, 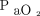 /
/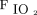 , etc.), most ICU clinicians would agree that such a patient is likely to be on the ventilator for > 7 d.41 Work in this area should focus on identifying the patients who are not expected to be on the ventilator very long but subsequently develop respiratory failure and require more long-term care. For example, further work in this area should help identify a medical patient sent to the ICU on a mechanical ventilator following a routine surgical procedure and is expected to be extubated within 24 h but develops significant respiratory failure that precludes the ICU team from being able to wean the ventilator rapidly, perform an extubation readiness assessment, and extubate the patient.
, etc.), most ICU clinicians would agree that such a patient is likely to be on the ventilator for > 7 d.41 Work in this area should focus on identifying the patients who are not expected to be on the ventilator very long but subsequently develop respiratory failure and require more long-term care. For example, further work in this area should help identify a medical patient sent to the ICU on a mechanical ventilator following a routine surgical procedure and is expected to be extubated within 24 h but develops significant respiratory failure that precludes the ICU team from being able to wean the ventilator rapidly, perform an extubation readiness assessment, and extubate the patient.
Weaning and Extubation
A hallmark of mechanical ventilation care is the implementation of an extubation readiness test and the associated practices. Despite a number of clinical efforts to improve the effectiveness of weaning and extubation practices, a number of challenges still remain.42 In a cohort of newborns, Mueller et al43 compared the performance of standard bedside practice with an artificial neural network (ANN) that incorporated 13 clinical parameters to classify extubation readiness. The ANN extubation model achieved an area under the receiver operating characteristic curve of 0.87. Importantly, this result was not vastly different from standard clinical practice. One interpretation of these results is that the ANN model is unnecessary. However, if an automated extubation classifier performed as well as standard care, it could free up clinicians to perform other tasks. After extending this work, the authors concluded that clinician predictions outperformed the ANN model.44 Hsieh et al45 conducted a similar study in an adult population and noted that performance was superior to the rapid shallow breathing index. Prasad et al46 proposed a data-driven approach to provide optimized mechanical ventilator weaning. They employed a Markov decision process to patient admissions to identify representations of patient condition, and they applied a reinforcement learning technique that learned a simple ventilator weaning protocol from historical data. Although a number of challenges are addressed in the paper, further work is needed to validate these results prospectively and to compare performance between the new extracted policy and standard practice properly.
Sepsis
Sepsis is a life-threatening condition that requires timely diagnosis and treatment to prevent tissue damage, organ failure, and death. Sepsis is the main cause of ARDS in approximately 70% of all cases and often requires mechanical ventilation.47 However, sepsis frequently is not recognized until later stages when symptoms have become severe.
Nemati et al48 analyzed a large cohort of subjects and trained a machine learning model that incorporated both demographic information as well as time-series features to predict sepsis risk 12 h before diagnosis. The area under the receiver operating characteristic curve was 0.83 (sensitivity 0.85, specificity 0.67). In a cohort of critically ill children, Kamaleswaren et al49 utilized a number of machine learning techniques (ie, logistic regression, random forests, deep convolution neural networks) to identify physiologic markers from time-series data to identify subjects with sepsis. Models utilizing convolution neural networks provided the best sensitivity and specificity (81% and 76%, respectively). Im-portantly, the authors provided some degree of explainability in their work and reported that the top features incorporated into the model were the standard deviation of diastolic blood pressure and average heart rate.
Predicting Mortality in the ICU
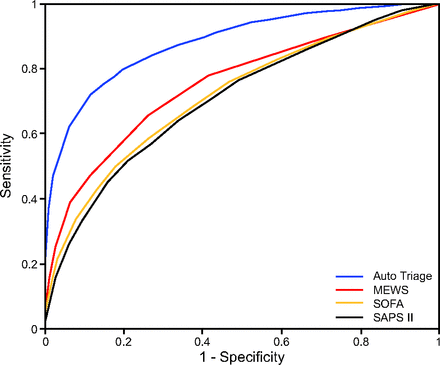
Calvert et al50 described a multi-dimensional analysis of clinical inputs to provide a mortality risk score for patients admitted to the ICU. Using only 8 common clinical factors found in the electronic health record, they reported that the results of their algorithm, AutoTriage, has an area under the receiver operating characteristic curve of 0.88 and a sensitivity of 80%. These results outperformed other common scores, including the Modified Early Warning Score (MEWS), Sequential Organ Failure Assessment (SOFA), and Simplified Acute Physiologic Score II (SAPS II) in a population derived from the MIMIC III database (Fig. 5).51,52
Tips on Getting Started
For clinicians and researchers unfamiliar with data science methods or machine learning, the task of starting a project can be daunting. Indeed, a significant activation energy is required to build up the required personnel, technical skills, and general best practice knowledge required to collect and clean data, train models, analyze performance, and draw sound conclusions based on results.
For those ready to learn a new language, you can download programming languages and get a large number of learning resources online at no cost. Two popular languages include Python (available at https://www.python.org, Accessed April 13, 2020) or R (available at https://www.r-project.org, Accessed April 13, 2020). Python is a free, high-level, general-purpose programming language with a large community support base that offers solutions for data wrangling, statistics, machine learning, and application development. R is a free statistical computing language and offers great support for managing large datasets, statistics, and machine learning. Matlab is a technical computing language that is available for a cost, but this is often available through an academic institution or the research computing department at many hospitals (see https://www.mathworks.com, Accessed April 13, 2020; for more information, contact your research support staff to inquire about licenses). Matlab offers a mature programming interface with many tasks that can be completed with the use of an easy graphical user interface; this software has good customer support, robust community forums, and is capable of tackling a wide variety of computing tasks, including statistics, machine learning, data visualization, and much more.
Although publications including machine learning methods are increasingly being accepted in mainstream clinical journals, the vast majority of papers exist in places a typical ICU clinician may not look. Rather than a simple https://PubMed.gov search (Accessed April 13, 2020), use resources like Web of Science (typically available through your institution) and https://scholar.google.com (Accessed April 13, 2020). These websites offer searches that include a number of journals outside those indexed in PubMed and can help identify important publications.
Ideally, one would already have a large data set of patients from his or her own institution that is ready for analysis. Indeed, the majority of the time dedicated to a project in the big data or artificial intelligence space will be committed to compiling, cleaning, and otherwise preparing your data for analysis. While this should be a priority, there are some high-quality data sets available to the public that can be used to get started. The Medical Information Mart for Intensive Care (MIMIC-III) is a freely accessible critical care database that contains > 60,000 ICU admissions and includes demographic and patient data (eg, laboratory, medications, and other health care data).53 The database was developed at the Massachusetts Institute of Technology Lab for Computational Physiology and can be accessed online at https://mimic.physionet.org (Ac-cessed April 13, 2020).
Future Direction
Management of the acutely ill patient in the ICU is not going to be completely automated anytime soon. As of this writing, no machine learning silver bullet exists that ingests data and spits out 100% accurate clinical predictions. Indeed, a number of publications discussed in this article offer levels of accuracy that aren’t terribly exciting; many papers report area under the receiver operating characteristic curve in the range of 0.7–0.8. In light of this, we may be tempted to conclude that artificial intelligence really doesn’t work in the ICU. However, this is more likely a function of limited data available to the model rather than a model’s inability to perform. It is essential to provide a comprehensive data set that contains as much information as possible to describe clinical conditions. If a data set contains variables that account for only 25% of the information available to clinicians, it should be no surprise that clinicians can make better predictions than the models. As ICUs increasingly implement systems that collect continuous data from mechanical ventilators and physiologic monitors, and transcribe laboratory data, imaging data, and information from clinical documentation in real time, we should see improvements in model performance as these factors are incorporated. Does this mean that all future studies should include machine learning? Certainly not. The simplest solution should always be preferred, and in many cases traditional analyses will suffice. However, applied machine learning, by nature of its ability to identify complex non-linear relationships among variables and to provide predictions of important clinical events, will be an important tool for those involved in research, quality improvement, and day-to-day ICU workflow.
Rather than simply making predictions with a machine learning model, future work should focus on offering decision support to bedside clinicians. For example, suppose that we want to develop a model to predict extubation success that won’t require respiratory therapists to manually conduct an extubation readiness test. Clinical data are recorded, and cases are labeled. Currently, many teams would attempt to build a prediction model that classifies a patient as simply successful or not successful. If that model doesn’t perform with tremendous accuracy, however, no reasonable clinician would trust it, and it would not be implemented. Rather, we should offer predictions that enable clinicians to incorporate clinical judgment. This way, the model will generate a prediction and label a patient as predicted to successfully extubate, but the model also will describe the probability of success (eg, 88% probability of successful extubation). Now a clinician is empowered. This model may identify that this patient is eligible for extubation earlier than is typically recognized. The clinical team can also decide if 88% probability is enough for this patient at this time. For low-risk patients who have simple intubations or who would likely tolerate noninvasive ventilation if needed, perhaps they can be extubated at this probability level. For higher-risk patients, such as those with a difficult airway or those who will not tolerate noninvasive ventilation, perhaps the care team should wait to extubate.
In general, artificial intelligence and big data are becoming an important part of the respiratory literature. As mature data-collection systems are implemented, these methods can be applied to build decision support tools to provide insight to bedside clinicians, but we still have a long way to go.
Discussion
Pham:
In terms of patient confidentiality and all these issues, is there a plan for international agreement of how we can obtain all these data coming from patients? I have the feeling that now there are so many wearables, the phone in your pocket, everybody has a lot of data on you but when you want to do research and collect it on patients it’s way more complicated to get patient or caregiver agreement for you to collect data. Even if it’s anonymous.
Smallwood:
That’s a good question. Just getting the data into your hands is important and sometimes difficult. One of the best things I think we can do to be successful is make the most of what you currently have. So rather than starting with a worldwide standard for all medical data we design a retrospective study at an individual ICU, perhaps grow that to partner with some ICUs who have access to similar data and go from there. That may take making very good friends with your information services department to get access to the SQL database where a lot of this information is buried. I know that in my institution we have a whole core of people who are trained in SQL to go in and work with me to extract relevant information that I need for a study. I’ve also managed to get myself access to the database for select projects. The IRB pathway to get to that is not too difficult either. I have to go through review with my department but once I do that it will be exempt from IRB because it’s all retrospective data, so there’s no real risk as long as I do a good job of protecting the private healthcare information. To your question about working through the pathway to obtain healthcare data, we certainly need to do a better job of connecting device data, medication administration record and other details from the patient’s chart all in one place. One of the things that’s difficult is mapping the conditions that are known to that patient at any given point in time because the coding used for billing is not optimized to patient care; it’s optimized for billing. I can extract an ICD-10 code for respiratory distress but if there’s some other code that will be more appropriate to get maximum reimbursement for that subject then I may miss some information. Combining it with other information, physiologic data, etc. can certainly start to solve the problem but I think in general we will see an increasing amount of ‘computable phenotypes’ for conditions we see in the ICU. Essentially some standard, reproducible method of obtaining information and assigning that patient some detail that’s important for both clinical and research purposes. There will be a whole host of problems moving from a purely retrospective exploration of information to then prospectively collecting data to validate, whether or not this will be reasonable to do in all cases is something to consider and may require changes to the way we consent people for care in the ICU is certainly on the table.
Goligher:
This is a really interesting and rapidly expanding area, like you pointed out and it’s really challenging to keep up so I really appreciate the way you spent time defining terms and clarifying the basic mechanics of machine learning, that was extremely helpful. The question I have for you is whether there’s any science that supports the notion that machine learning actually improves our ability to predict over conventional regression models? I’ve attempted machine learning studies 3 times now and in all 3 cases we were not able to improve our predictive performance with these very advanced machine learning techniques. It’s disappointing; obviously it’s not a magic bullet if you don’t have the information you need, you don’t have the information you need. But I wonder if you know of a systematic review or something like that that identified a proportion of cases where these new techniques really do improve our ability to discriminate outcomes of interest?
Smallwood:
I think that is one of the most important points of adopting this within the medical research community. I did an experiment myself, I said, ‘great, neural networks sound awesome, they’re going to learn better than I can, identifying things I never knew existed and essentially all of our problems.’ What I decided to do was predict carbon dioxide elimination and energy expenditure in a cohort of critically ill mechanically ventilated subjects. I cleaned up the data, I had something like 5 or so predictor variables, a good size population and I set to work training, optimizing, adjusting hyper-parameters, re-training and re-optimizing a neural network. It did horrible. What I found was that I could do a much better job combining what I believed to be important about the patients, what I knew about physiology and ended up with a much better prediction model. What I learned from that is that it’s not as simple as saying machine learning is better. It’s really about adding these skills to our toolbox, and recognizing when and how they can offer us superior insights or prediction performance in certain cases. By the way, a computer scientist would point out linear regression is technically machine learning. But take linear regression for example. You conduct an experiment and you want to describe the relationship between variable X and measurement Y. You apply linear regression and you don’t get a good R2 value. Do you blame linear regression for that? No. You’d simply conclude that the data don’t fit that model and you keep hunting for a better one. You can imagine many cases where that would be the case. I think that one of the things that happens to machine learning is that it’s good at identifying non-linear patterns in data. But in order for it to learn it needs a heck of a lot of data, and a lot more than is typically collected for something like a pilot study. The sheer volume of data required to actually make the thing work how it’s supposed to can be beyond what’s practical for a single institution. That’s certainly not always the case but something to watch out for.
Lamberti:
I would refer you to a recent study from the University of Chicago that utilized machine learning to improve the clinical prediction of death, cardiac arrest or ICU transfer in general care unit patients.1 A standard regression analysis using vital signs to determine the risk of clinical deterioration yielded an area under the receiver operating characteristic curve (AUC) of 0.74. They utilized machine learning (random forest) and were then able to improve the AUC to 0.80. Machine learning did improve the clinical predictive value. But, is changing the AUC by 0.06 clinically meaningful?
Smallwood:
There’s also a review in NEJM2 that went through, not necessarily the whole spectrum of machine learning, but the general ways we think about problems and some of the limitations of machine learning.
Schmidt:
Suppose you had a really good predictive tool, have you thought about some of the downstream challenges who will see that, who will act on that, how will that be integrated with the current models of care provision?
Smallwood:
That’s a good question: we have something great but we can’t use it. I think that will be a real problem as we move forward. I think most ICUs haven’t approached that problem yet but we are certainly getting there. Like any other piece of technology that we purchase at a hospital, the adoption and utilization need to be the first thing we think about. We can buy a really excellent monitor that has the capacity to apply advanced predictive modelling for our patients, but if people aren’t educated, don’t trust it and therefore don’t actually use it, or if the alerts aren’t helpful to the clinicians, then it’s all for naught. Your point is very well taken, that a lot more care will have to be taken around the actual implementation science of the decision support model itself and not just making a really great tool. This will certainly require high level buy-in at the hospital to set up the computing infrastructure required to collect, analyze and provide decision support alerts and that’s definitely a touch challenge, but one I believe can be eventually justified.
Goligher:
One of the things that’s an important limitation of this whole business is the fact that you can retrospectively, particularly with unsupervised techniques, identify clusters of patients. The kind of paradigm I’m thinking of is Calfee’s seminal work on subphenotypes of ARDS.3-5 And every time they go to a new dataset they’re able to identify these subphenotypes which consistently look like hyper- and hypo-inflammatory subphenotypes. The problem is they can’t actually go to the emergency department and look at a patient who’s newly diagnosed with ARDS and decide which group they’re in. There’s no way to prospectively classify patients, it’s always a retrospective unsupervised analysis of previously collected data. In order to try and actually overcome that problem you still have to have theory-driven hypothesis-based tests that are thinking about, ‘what basic mechanisms would drive a patient being in one group or the other?’ or try to develop prospective ways of classifying patients. It seems to me like we can identify subgroups but then the challenge of making that useful prospectively in relation to Craig’s point remains an important problem to be surmounted.
Smallwood:
That’s very well said and I think something that will have to be mostly solved for different applications. In general, any clustering algorithm is going to be most helpful retrospectively and can be hypothesis generating. On the other hand, a number of regression machine learning models are more built to make predictions and would be better suited to offer bedside decision support. In the context of ARDS sub-phenotypes and classifying patients in near-real time. The next step may be to design a study where those groups identified from clustering are labeled 1 and 2. Take some clinical data available at the time of admission in the ER and apply a regression algorithm like a support vector machine and try to predict either class 1 or class 2. In that way the project evolves from identifying groups that weren’t clinically obvious, noting some important clinical reason why they may respond better or worse to treatment, then predicting that class with available data. But as some stage it’s going to come down to designing your whole data infrastructure around the clinical application of these models, because up until this point we haven’t really designed our monitors, our information systems, our EHR around getting information out, churning it, and then spitting out a helpful recommendation. That’s just not how it was designed. In some data scientist’s office, he or she can spend months doing retrospective analysis and you can let a computer look at a single patient for a number of hours and it doesn’t matter. There’s no rush. But in the ICU we need to be much quicker than that. One of the things I’m trying to work on at my institution is designing, at least in the ICU, a mechanism that enables information to be available to a number of applications, whether it be for research, third party applications, custom built algorithms, etc. We need a way to spin that information back to the clinician at the bedside. I feel the pain of seeing a good result in a paper and having no way to make that a reality at the bedside.
Blanch:
All of this to me should be done according to a hypothesis you’re trying to solve. And then you create the tools to answer the question. We realize that, for example, in waveforms there are events that happen which might be important but still not diagnosed. Using supervised machine learning, physiologic knowledge behind is needed. Information in large ICU databases is complex and might be incomplete or corrupted and needs supervision, modeling to improve quality. It’s an enormous problem like you said.
Smallwood:
You touched on a couple of good points, Lluís. One of which is the hype cycle. We see this all the time and not just with medical technology. It goes something like this. New technology gets introduced and there is a great deal of excitement, ‘this new technology will solve all of our problems!’ Well, it doesn’t. We need to understand it, have a solid understanding of the problem we have and implement this new technology in thoughtful and systematic ways. I know that my presentation and the forthcoming paper sounds like I’m a big data and machine learning zealot, but frankly I’m not and I find myself mostly agreeing with your point of view Dr. Blanch. However, I do believe that machine learning, along with the clinicians and invested and knowledgeable researchers will actually solve a number of important problems. That said, we should always prefer the simpler solution to a problem rather than a complicated one. In many cases we may not need sophisticated algorithms at all and that is completely appropriate. But in a number of cases I think we will find that the standard methods that all of us have seen in the medical literature for decades will be found lacking and we will do well to have big data and machine learning skills available to us. One opportunity I believe we have is to do a better job of identifying problems based on outcome data. Indeed, I struggle with myself. Often, the way we select an area to study is based on personal interest or maybe your chief or medical director says we need to work on this problem. A lot of time that may work out just fine. But what if our areas of improvement were data driven and based on automated outcome assessment? I think an emerging area will be the combination of automated outcome assessment, facilitated with big data methodologies in order to identify areas of our practice that need improvement. That will include taking into account how big of an effect it will have on the patients morbidity and mortality, cost of care, but also how practical our envisioned solution is to make happen at the bedside.
Walsh:
Craig, would you share with us, in your ideal world where everybody is connected and the data are clean, how could you potentially take advantage of the actual experiments that are happening daily we call practice of medicine? We have modes of ventilation, all of us have biases of things we think are important and less important. If we were able to get all of that data could you share with us what that would mean?
Smallwood:
I’ll tell a story that illustrates this. I work in a pediatric ICU. I see mortality of ARDS as relatively low compared to adults. If you look at the literature it’s about 18%. If you ask the simple question, at any given moment in time, what is our ICU mortality? And how can we benchmark against everybody else? There’s no easy way to do that. In a single institution, there are different databases, each put together for different reasons but collect some of the same data. In one data base you may a 33% mortality. If I trust that number, then alarm bells better be going off because on the surface that is much worse than what is expected. Of course, there are a number of things we need to assess the under-diagnosis of mild and moderate ARDS, the severity of illness and some other factors that would change how I interpret comparing those 2 numbers. So #1, can I trust that information, and #2 if I can then I go into the data and explore what perhaps led to those outcomes. I think that that exploration isn’t necessarily machine learning it’s more datamining but essential, nonetheless. But to your question Brian, let’s suppose we have this all figured out, we trust the data, a number of different institutions implement the same methodologies. What’s next? What I would love to see is identifying cohorts of patients who had a favorable outcome and exploring factors associated with that. In some cases, it may just be severity of disease, or some hidden phenotype that was previously unrecognized. But what if we spot some differences in care? I think that would be hypothesis-generating. And the beauty of this approach is that these hypotheses may not have been obvious before going through this exercise. This could inform the development of algorithms based on better outcomes and utilized prospectively. Of course, prospective evaluation will be important before proving to ourselves that we have actually improved care. In general, I think this should be easy one day. Right now, it’s not. But that’s what I’d like to see going forward.
Blanch:
Another thing, in the past the relationships between heath institutions, academia and industry were reasonably established. But now seems there is some overlap and big players like Apple or Google are new actors. I’d like your opinion on how should we work or regulated intellectual property with these big players.
Smallwood:
I think it really comes down to how in the boundaries of the law, we can develop improvements in patient care. In the past there was this whole mechanism, how to better build a sensitivity mechanism to make NIV triggering better for patients. There’s no mechanism for me at my institution to build that and put it on our patients. What you have to do is partner closely with the developer or the company that made that device, go through a whole pathway of validation experiments, go through a whole regulatory pathway to get it approved, and then actually apply it to patients in the ICU. What’s nice about that is the whole collaboration is where the lines are drawn. But when we talk about machine learning we do all kinds of things for clinical improvements and changes in care that don’t require us to go through the FDA. And where does some model that tells me to change or do something different with a mechanically ventilated patient stay? Is it okay for me to look at my own information at my hospital, optimize some model and then push it as a clinical initiative? Do I even need to go to the FDA? If I do, then you can consider who to partner with and rapidly turn that around the whole regulatory pathway. But it’s not necessarily clear to me that would be the case 100% of the time. What I would love to see is a mechanism at individual institutions where we have a group of people who ensure that the algorithm has demonstrated reliability, accuracy of data and data showing it’s superiority to current clinical practice. This wouldn’t be that different from a scientific review board, or a quality committee evaluating a new guideline. The key difference will be the additional knowledge required to ask informed questions about the technical performance of the algorithm, data reliability, etc. Those skills are currently outside the average quality improvement committee. There may be cases where that’s not appropriate and a broader federal regulation will be required but in general that’s what I think we will see moving forward. But regulation usually follows innovation so we will have to see what happens.
Footnotes
A version of this paper was presented at the 58th Respiratory Care Journal Conference, held June 10–11, 2019, in St Petersburg, Florida.
Dr Smallwood has disclosed a relationship with Capsule Technologies.
- Copyright © 2020 by Daedalus Enterprises